With four interactive research work packages ImageInLife aims to bring major improvements at the level of the main bottlenecks in the field of vertebrate imaging.
The goal is to reach cellular or subcellular resolution within a complete organism. This is now possible using vertebrate embryos as models and the most recent technological advances as tools. Through mutualisation and standardisation of protocols, the ultimate scientific aim and research objective of the network is to provide a common toolbox (markers, microscope setups and software) to the scientific community. This toolbox is necessary to reach a level of phenomenological and quantitative description allowing a better understanding of the developmental and pathological processes within the vertebrate embryo. It can also be implemented in other research fields or applications directed toward growing international markets (microscopes, screenings and medical imaging).
Research work packages (WP) of ImageInLife:
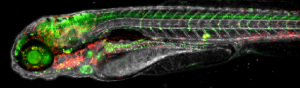
WP1: Multilevel imaging of vertebrate development and disease.
Finding the right conditions for optimal imaging of embryos from different vertebrate species (zebrafish, mice & rabbit). Conditions will have to be improved both to access developmental stages that are not currently available due to the intra-utero development of rabbit and mouse embryos and to obtain long imaging sessions of developing embryos whose shape is constantly changing. In order to study developmental processes and pathologies at the cellular or subcellular level, new markers and sensors must be developed for optimal imaging (membrane and nucleus labelling and new colour combinations). New probes and sensors will have to be compatible with both the biological material and its incubation conditions and with the microscopes to be developed within WP2. This WP1 should lead to new solutions for companies (Acquifer, P9) using the vertebrate embryo for High Content Screening (HCS).
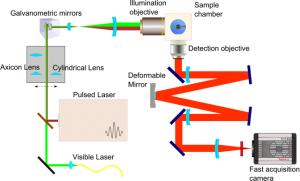
WP2: Tools and instrumentation for multiscale in vivo imaging.
Bringing small developing embryos to the microscope and keeping them as healthy as possible during long imaging sessions is not an easy task. This step has to be very innovative since optimal incubation chambers must still be developed, which will require an excellent knowledge of the biological material, mastery of the optical requirements of the chosen microscopy technologies and a good knowledge of microfluidics. Research efforts will be dedicated to adapting new microscopy solutions to the vertebrate embryos: high resolution and speed for transmission imaging, light sheets (structured or not) and single molecule fluorescent imaging. The aim is to design a versatile optical setup allowing imaging at different magnifications and resolutions, with different sheets, beam profiles (Gaussian and Bessel) and regimes (linear and nonlinear) with several imaging paths for multiview imaging and new cameras (sCMOS for 3D volume acquisition at camera frame rate). It will also include adaptive optics, beam shaping, confocal line scan strategies and remote focussing. Super-resolution strategies will be correlated with light-sheet based in vivo study of biological processes within the zebrafish embryo. All these issues are challenging avenues to be explored and are closely backed by companies that could bring new solutions to the market.
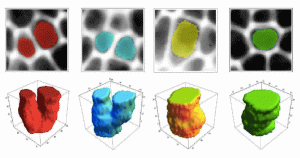
WP3: Image processing and analysis.
The aim of WP3 is to explore and build new mathematical schemes and computing strategies for segmentation and tracking in 3D and 4D biomedical images produced by WP1&2. For cell tracking, the issue is to deal with the spatiotemporal nature of the data and the level of noise. Segmentation is critical both for cells and subcellular elements to be imaged, but also at a lower resolution for tissue and higher order organisation patterning. These new solutions for segmentation and cell tracking will lead, through analogies, to new solutions for the analysis of medical images.
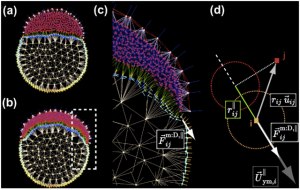
WP4: Image-based integrative and predictive modelling of complex biological processes.
Based on the reconstructed data (WP3) measured from microscopy imaging, the objectives of WP4 will be to understand and predict the structure and function of biological systems via computational and mathematical modelling, followed by a systematic and evolutionary exploration of parameter space to optimise the “fitness” of virtual embryos compared to real data.